Online first
Current issue
Archive
Special Issues
About the Journal
Publication Ethics
Anti-Plagiarism system
Instructions for Authors
Instructions for Reviewers
Editorial Board
Editorial Office
Contact
Reviewers
All Reviewers
2025
2024
2023
2022
2021
2020
2019
2018
2017
2016
General Data Protection Regulation (RODO)
RESEARCH PAPER
Investigation for the presence of bacteria and antimicrobial resistance genes in sea snails (Rapana venosa)
1
Department of Medical Services and Techniques, Vocational School of Health Services, Firat University, Elazig, Turkey
2
Department of Medical Microbiology, Faculty of Medicine, İnönü University, Malatya, Turkey
3
Department of Aquaculture and Fish Diseases, Fisheries Faculty, Firat University, Elazig, Turkey
4
Center for Immunology and Infectious Diseases, University of California, Davis (CA), USA
5
Department of Animal Nutrition and Husbandry, University of Veterinary Medicine and Pharmacy, Košice, Slovak Republic
Corresponding author
František Zigo
University of Veterinary Medicine and Pharmacy in Košice, UVLF, Komenského 73, Košice, Slovakia, 04001, Košice, Slovak Republic
University of Veterinary Medicine and Pharmacy in Košice, UVLF, Komenského 73, Košice, Slovakia, 04001, Košice, Slovak Republic
Ann Agric Environ Med. 2023;30(2):235-243
KEYWORDS
TOPICS
- Biological agents posing occupational risk in agriculture, forestry, food industry and wood industry and diseases caused by these agents (zoonoses, allergic and immunotoxic diseases)
- Health effects of chemical pollutants in agricultural areas , including occupational and non-occupational effects of agricultural chemicals (pesticides, fertilizers) and effects of industrial disposal (heavy metals, sulphur, etc.) contaminating the atmosphere, soil and water
ABSTRACT
Introduction and objective:
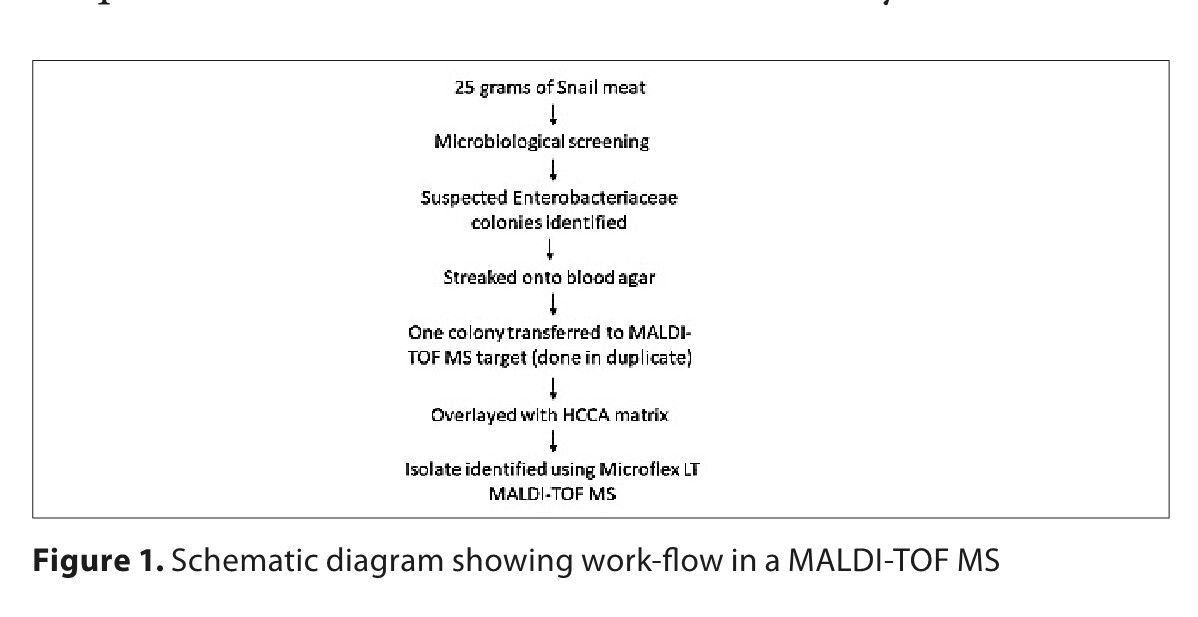
The aims of this study were to search for the presence of bacteria in sea snails (Rapana venosa) by using culturomics and Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS), and the antibiotic resistance/susceptibility of the sea snails.
Material and methods:
The anti-microbial susceptibilities of Gram-negative bacteriawas assessed by the Kirby-Bauer disk diffusion method, the presence of the mcr genes (mcr-1 to -5), the major carbapenemase and β-lactamase resistant genes in Gram-negative bacteria, using mPCR method and 16S rRNA sequence analysis of A. hydrophila isolates.
Results:
Bacterial growth accounted for 100% and 94.2% in the samples of intestine and meat, respectively, in the snails. The main organisms identified by MALDI-TOF MS were A. salmonicida subsp. salmonicida at 33.7%, followed by Raoultella ornithinolytica at 9.6% (10/104) and Staphylococcus warneri at 7.7% in meat and intestine samples. Aeromonas hydrophila/punctata (caviae), Aeromonas sobria, Klebsiella aerogenes, Klebsiella oxytoca, Raoultella planticola, Shewanella putrefaciens and Vibrio vulnificus are intrinsic or chromosomally-mediated resistant against ampicillin. No mcr genes (mcr-1 to -5), the major carbapenemase and β-lactamase resistant genes were found. Aeromonas salmonicida subsp. salmonicida showed very low levofloxacin and meropenem resistance levels at 2.9%. When the sequence was searched in the Blast database, the genome of A. hydrophila/punctata (caviae) isolate showed high similarity with the A. hydrophila sequences.
Conclusions:
Conclusions. The findings obtained not only provide data about the proportion of bacteria in the gut and meat of the sea snails and their antibiotic resistance/susceptibility, but also show the absence of carbapenemase, colistin, and β-lactamase resistant genes among bacterial isolates from sea snail gut microbes.
The aims of this study were to search for the presence of bacteria in sea snails (Rapana venosa) by using culturomics and Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS), and the antibiotic resistance/susceptibility of the sea snails.
Material and methods:
The anti-microbial susceptibilities of Gram-negative bacteriawas assessed by the Kirby-Bauer disk diffusion method, the presence of the mcr genes (mcr-1 to -5), the major carbapenemase and β-lactamase resistant genes in Gram-negative bacteria, using mPCR method and 16S rRNA sequence analysis of A. hydrophila isolates.
Results:
Bacterial growth accounted for 100% and 94.2% in the samples of intestine and meat, respectively, in the snails. The main organisms identified by MALDI-TOF MS were A. salmonicida subsp. salmonicida at 33.7%, followed by Raoultella ornithinolytica at 9.6% (10/104) and Staphylococcus warneri at 7.7% in meat and intestine samples. Aeromonas hydrophila/punctata (caviae), Aeromonas sobria, Klebsiella aerogenes, Klebsiella oxytoca, Raoultella planticola, Shewanella putrefaciens and Vibrio vulnificus are intrinsic or chromosomally-mediated resistant against ampicillin. No mcr genes (mcr-1 to -5), the major carbapenemase and β-lactamase resistant genes were found. Aeromonas salmonicida subsp. salmonicida showed very low levofloxacin and meropenem resistance levels at 2.9%. When the sequence was searched in the Blast database, the genome of A. hydrophila/punctata (caviae) isolate showed high similarity with the A. hydrophila sequences.
Conclusions:
Conclusions. The findings obtained not only provide data about the proportion of bacteria in the gut and meat of the sea snails and their antibiotic resistance/susceptibility, but also show the absence of carbapenemase, colistin, and β-lactamase resistant genes among bacterial isolates from sea snail gut microbes.
ACKNOWLEDGEMENTS
The study was supported by a KEGA grant No. 006UVLF-
4/2020.
REFERENCES (77)
1.
Dar MA, Pawar KD, Pandit RS. Gut microbiome analysis of snails: A Biotechnological Approach. In: Ray S, editor. Organismal and Molecular Malacology. Rijeka: InTech; 2017. p. 189–217.
2.
Terzi E. Antimicrobial resistance profiles and tetracycline tesistance genes of Escherichia coli in Mediterranean mussel and sea snails collected from the Eastern Black Sea (Turkey). Alinteri J Agric Sci. 2018;33:43–49.
3.
Civelek F. Determination of heavy metal resistance levels in Escherichia coli isolated from Mediterranean mussel and sea snail in Eastern Black Sea. Recep Tayyip Erdogan University, Graduate School of Natural and Applied Sciences, Department of Fisheries, Master Thesis; 2019.
4.
Pissia M?, Matsakidou A, Kiosseoglou V. Raw materials from snails for food preparation. Future Foods. 2021;3:100034. https://doi.org/10.1016/j.fufo....
5.
Song H, Yu ZL, Yang MJ, et al. Analysis of microbial abundance and community composition in esophagus and intestinal tract of wild veined rapa whelk (Rapana venosa) by 16S rRNA gene sequencing. J General Applied Mic. 2018;64:158–166.
6.
Eichenberger EM, Thaden JT. Epidemiology and mechanisms of resistance of extensively drug resistant Gram-negative bacteria. Antibiotics (Basel). 2019;8:37. https://doi.org/10.3390/antibi....
7.
Poirel L, Jayol A, Nordmann P. Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clinical Mic Rev. 2017;30:557–596.
8.
Liu YY, Wang Y, Walsh TR, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Inf. 2016;2:161–8. https://doi.org/10.1016/s1473-....
9.
Xavier BB, Lammens C, Ruhal R, et al. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium. Euro Surv. 2016;21:27. https://doi.org/10.2807/1560-7....
10.
Yin W, Li H, Shen Y, et al. Novel plasmid-mediated colistin resistance gene mcr-3 in Escherichia coli. mBio. 2017;8:e00543–17. https://doi.org/10.1128/mbio.0....
11.
Carretto E, Brovarone F, Nardini P, et al. Detection of mcr-4 positive Salmonella enterica serovar Typhimurium in clinical isolates of human origin, Italy, October to November 2016. Euro Surv. 2018;23:17–21.
12.
Borowiak M, Fischer J, Hammerl JA, et al. Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi B. J Antimicrobial Chem. 2017;72:3317–3324.
13.
AbuOun M, Stubberfield EJ, Duggett NA, et al. mcr-1 and mcr2 (mcr-6.1) variant genes identified in Moraxella species isolated from pigs in Great Britain from 2014 to 2015. J Antimicrob Chem. 2018;73(10):2904. https://doi.org/10.1093/jac/dk....
14.
Yang YQ, Li YX, Lei CW, et al. Novel plasmid-mediated colistin resistance gene mcr-7.1 in Klebsiella pneumoniae. J Antimicrobial Chem. 2018;73:1791–1795.
15.
Wang R, van Dorp L, Shaw LP, et al. The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nature Commun. 2018;9:1179. https://doi.org/10.1038/s41467....
16.
Carroll LM, Gaballa A, Guldimann C, et al. Identification of novel mobilized colistin resistance gene mcr-9 in a multidrug-resistant, colistin-susceptible Salmonella enterica serotype Typhimurium isolate. mBio, 2019;10:e00853–19. https://doi.org/10.1128/mbio.0....
17.
Wang C, Feng Y, Liu L, et al. Identification of novel mobile colistin resistance gene mcr-10. Emer Microbes Inf. 2020;9:508–516.
18.
Luo Q, Wang Y, Xiao Y. Prevalence and transmission of mobilized colistin resistance (mcr) gene in bacteria common to animals and humans. Biosaf Health. 2020;2:71–78. https://doi.org/10.1016/j.bshe....
19.
Falagas ME, Kasiakou SK. Colistin: the revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin Inf Diseases. 2005;40:1333–1341.
20.
Otlu B, Yakupogullari Y, Gürsoy NC, et al. Co-production of OXA-48 and NDM-1 carbapenemases in Providencia rettgeri: the first report. Mikrobiyoloji Bulteni. 2018;52:300–307.
21.
Gurung S, Kafle S, Dhungel B, et al. Detection of OXA-48 gene in carbapenem-resistant Escherichia coli and Klebsiella pneumoniae from urine samples. Inf Drug Res. 2020;13:2311–2321.
22.
Queenan AM, Bush K. Carbapenemases: The versatile ß-lactamases. Clin Mic Rev. 2007;20:440–458.
23.
Ejaz H, Younas S, Abosalif KOA, et al. Molecular analysis of blaSHV, blaTEM, and blaCTX-M in extended-spectrum ß-lactamase producing Enterobacteriaceae recovered from fecal specimens of animals. PLoS One. 2021;16:e0245126. https://doi.org/10.1371/journa....
24.
Jouini A, Vinué L, Slama K, et al. Characterization of CTX-M and SHV extended-spectrum beta-lactamases and associated resistance genes in Escherichia coli strains of food samples in Tunisia. J Antimicrobial Chem. 2007;60:1137–1141.
25.
Gundran RS, Cardenio PA, Villanueva MA, et al. Prevalence and distribution of bla CTX-M , bla SHV , bla TEM genes in extended-spectrum ß-lactamase-producing E. coli isolates from broiler farms in the Philippines. BMC Vet Res. 2019;15:227.
26.
Bajpai T, Pandey M, Varma M, Bhatambare GS. Prevalence of TEM, SHV, and CTX-M beta-lactamase genes in the urinary isolates of a tertiary care hospital. Avicenna J Med. 2017;7:12–16.
27.
Shen Y, Yin W, Liu D, et al. Reply to Cabello, et al. Aquaculture and mcr colistin resistance determinants. mBio. 2018a;3(5):e01229–17. https://doi.org/10.1128/mbio.0....
28.
Shen Y, Zhou H, Xu J, et al. Anthropogenic and environmental factors associated with high incidence of mcr-1 carriage in humans across China. Nature Mic. 2018b;3:1054–1062.
29.
Altug G, Güler N. Determination of the levels of indicator bacteria, Salmonella spp. and heavy metals in sea snails (Rapana venosa) from the Northern Marmara Sea, Turkey. Turkish J Fisheries Aquatic Sci. 2002;2:141–144.
30.
Irkin R, Korukluoglu M, Tavşanli H. Microbial properties of some sea products intended for export. Türk Hijyen ve Deneysel Biyoloji Dergisi. 2007;64:26–31.
31.
Florio W, Baldeschi L, Rizzato C, et al. Detection of Antibiotic-Resistance by MALDI-TOF Mass Spectrometry: An Expanding Area. Front Cell Infect Microbiol. 2020;10:572909.
32.
Flores-Trevino S, Garza-Gonzalez E, Mendoza-Olazaran S, et al. Screening of biomarkers of drug resistance or virulence in ESCAPE pathogens by MALDI-TOF mass spectrometry. Sci Rep. 2019;9:18945.
33.
Patel R. Matrix-assisted laser desorption ionization-time of flight mass spectrometry in clinical microbiology. Clin Infect Dis. 2013;57:564–572.
34.
Wang Y, Chen XF, Xie XL, et al. Evaluation of VITEK MS, Clin-ToF-II MS, Autof MS 1000 and VITEK 2 ANC card for identification of Bacteroides fragilis group isolates and antimicrobial susceptibilities of these isolates in a Chinese university hospital. J Microbiol Immunol Infect. 2019;52:456–464.
35.
Barnini S, Ghelardi E, Brucculeri V, Morici P, Lupetti A. Rapid and reliable identification of Gram-negative bacteria and Gram-positive cocci by deposition of bacteria harvested from blood cultures onto the MALDI-TOF plate. BMC Microbiol. 2015;15:124.
36.
Bryson AL, Hill EM, Doern CD. Matrix-Assisted Laser Desorption/Ionization Time-of-Flight: The Revolution in Progress. Clin Lab Med. 2019;39:391–404.
37.
Oviano M, Sparbier K, Barba MJ, Kostrzewa M, Bou G. Universal protocol for the rapid automated detection of carbapenem-resistant Gram-negative bacilli directly from blood cultures by matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry (MALDI-TOF/MS). Int J Antimicrob Agents. 2016;48:655–660.
38.
Pupo GM, Lan R, Reeves PR. Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc Natl Acad Sci. USA. 2000;97:10567–10572.
39.
Fangous MS, Mougari F, Gouriou S, et al. Classification algorithm for subspecies identification within the Mycobacterium abscessus species, based on matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2014;52:3362–3369.
40.
Andrews WH, Jacobson A, Hammack T. Salmonella, chapter 5. In: Hammack T, Feng P, Jinneman K, et al. Bacterial analytical manual. Silver Spring: Food and Drug Admin; 2011. p. 276.
41.
Elhadi N, Prevalence and antimicrobial resistance of Salmonella spp. in raw retail frozen imported freshwater fish to Eastern Province of Saudi Arabia. Asian Pacific J Tropical Biom. 2014;4:234–238.
42.
Dubois D, Grare M, Prere MF, et al. Performances of the Vitek MS matrix-assisted laser desorption ionization-time of flight mass spectrometry system for rapid identification of bacteria in routine clinical microbiology. J Clinical Mic. 2012;50:2568–2576.
43.
Westblade LF, Jennemann R, Branda JA, et al. Multicenter study evaluating the Vitek MS system for identification of medically important yeasts. J Clinical Mic. 2013;51:2267–2272.
44.
Ellington MJ, Kistler J, Livermore DM, Woodford N. Multiplex PCR for rapid detection of genes encoding acquired metallo-beta-lactamases. J Antimicrobial Chem. 2007;59:321–322.
45.
Hasman H, Mevius D, Veldman K, et al. Beta-Lactamases among extended-spectrum beta-lactamase (ESBL)-resistant Salmonella from poultry, poultry products and human patients in the Netherlands. J Antimicrobial Chem. 2005;56:115–121.
46.
Poirel L, Heritier C, Tolun V, Nordmann P. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrobial Agents Chem. 2004;48:15–22.
47.
McCabe KM, Zhang YH, Huang BL, et al. Bacterial species identification after DNA amplification with a universal primer pair. Mol Genetics Met. 1999;66:205–211.
48.
Rebelo AR, Bortolaia V, Kjeldgaard JS, et al. Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Euro Surveill. 2018;23:17–00672.
49.
Poirel L, Dortet L, Bernabeu S, Nordmann P. Genetic features of blaNDM-1positive Enterobacteriaceae. Antimicrobial Agents Chem. 2011;55:5403–5407.
50.
European Committee on Antimicrobial Susceptibility Testing (EUCAST). Breakpoint Tables for Interpretation of MICs and Zone Diameters; EUCAST, Version 11.1. 2021.
51.
Hassan J, El-Gemayel L, Bashour I, Kassem II. On the edge of a precipice: The global emergence and dissemination of plasmid-borne mcr genes that confer resistance to colistin, a last-resort antibiotic. In: Hashmi MZ, editor. Antibiotics and Antimicrobial Resistance Genes in the Environment. Amsterdam, The Netherlands: Elsevier BV; 2020. p. 155–182.
52.
Joynson R, Pritchard L, Osemwekha E, Ferry N. Metagenomic analysis of the gut microbiome of the common black slug Arion ater in search of novel lignocellulose degrading enzymes. Frontiers Mic. 2017;8:2181.
53.
Charrier M, Fonty G, Gaillard-MartinIe B, et al. Isolation and characterization of cultivable fermentative bacteria from the intestine of two edible snails, Helix pomatia and Cornuaspersum (Gastropoda: Pulmonata). Biological Res. 2006;39:669–681.
54.
Hu Z, Tong, Q, Chang J, et al. Gut bacterial communities in the freshwater snail Planorbella trivolvis and their modification by a non-herbivorous diet. Peer J. 2021;9:e10716. https://doi.org/10.7717/peerj.....
55.
Janda JM, Abbott SL. The genus Aeromonas: Taxonomy, pathogenicity, and infection. Clinical Mic Rev. 2010;23:35–73.
56.
Salvat MJF, Ashbolt N. Global Water Pathogen Project. University of Alberta; Edmonton, AB, Canada. 2019. https://en.unesco.org/theme/wa....
57.
Figueras MJ, Beaz-Hidalgo R. Aeromonas. Academic Press; Norfolk, UK: Aeromonas infections in humans. 2015; 65–108.
58.
Latif-Eugenin FL, Aeromonas, un microorganismo ambiental de importancia en salud humana y animal. Universidad Rovira i Virgili; Tarragona, Spain. 2015; 406.
59.
Vincent AT, Fernández-Bravo A, Sanchis M, et al. Investigation of the virulence and genomics of Aeromonas salmonicida strains isolated from human patients. Inf Genetics Evol. 2019;68:1–9.
60.
Castro-Escarpulli G, Figueras MJ, Aguilera-Arreola G, et al. Characterisation of Aeromonas spp. isolated from frozen fish intended for human consumption in Mexico. Int J Food Microbiol. 2003;84:41–49.
61.
Figueras MJ, Beaz-Hidalgo R. Aeromonas: detection by cultural and modern techniques. Batt C, Tortorello ML, editor. Encyclopedia of food microbiology. Oxford, UK: Elsevier Ltd; 2014. p. 25–30.
62.
Woodring J, Srijan A, Puripunyakom P, et al. Prevalence and antimicrobial susceptibilities of Vibrio, Salmonella, and Aeromonas isolates from various uncooked seafoods in Thailand. J Food Protection . 2012;75:41–47.
63.
Yano Y, Hamano K, Tsutsui I, et al. Occurrence, molecular characterization, and antimicrobial susceptibility of Aeromonas spp. in marine species of shrimps cultured at inland low salinity ponds. Food Mic. 2015;47:21–27.
64.
Popović NT, Kazazić SP, Strunjak-Perović I, Čož-Rakovac R. Differentiation of environmental aquatic bacterial isolates by MALDI-TOF MS. Environmental Res. 2017;152:7–16.
65.
Virsek MK, Lovsin MN, Koren S, et al. Microplastics as a vector for the transport of the bacterial fish pathogen species Aeromonas salmonicida. Marine Poll Bulletin. 2017;125:301–309.
66.
Samasti M, Koçoglu ME, Davarci I, et al. Investigation of carbapenemase genes and clonal relationship in carbapenem resistant Klebsiella pneumoniae strains. Bezmialem Sci. 2019;7:186–190.
67.
Kuskucu MA, Karakullukcu A, Ailiken M, et al. Investigation of carbapenem resistance and the first identification of Klebsiella pneumoniae carbapenemase (KPC) enzyme among Escherichia coli isolates in Turkey: A prospective study. Travel Med Inf Dis. 2016;14:572–576.
68.
Codjoe FS. Detection and characterisation of carbapenem-resistant gram negative bacilli infections in Ghana. Doctoral, Sheffield Hallam University. 2016. Thesis (Doctoral). http://shura.shu.ac.uk/id/epri....
69.
Patel JB, Rasheed JK, Kitchel B. Carbapenemases in Enterobacteriaceae: Activity, epidemiology and laboratory detection. Clin Microbiol Newsletter. 2009;31:55–62.
70.
Cohen Stuart J, Leverstein-Van HMA. Dutch Working Party on the Detection of Highly Resistant Microorganisms. Guideline for phenotypic screening and confirmation of carbapenemases in Enterobacteriaceae. Int J Antimicrobial Agents. 2010;36:205–210.
71.
Codjoe FS, Donkor ES. Carbapenem resistance: A review. Medical Sci (Basel), Switzerland). 2017;6(1):E1. https://doi.org/10.3390/medsci....
72.
Abbott I, Cerqueira GM, Bhuiyan S, Peleg AY. Carbapenem resistance in Acinetobacter baumannii: Laboratory challenges, mechanistic insights and therapeutic strategies. Expert Rev Anti-infective Therapy. 2013;11:395–409.
73.
Woodford N, Turton JF, Livermore DM. Multiresistant Gram-negative bacteria: The role of high-risk clones in the dissemination of antibiotic resistance. FEMS Mic Rev. 2011;35:736–755.
74.
Grevskott DH, Svanevik CS, Sunde M, et al. Marine bivalve mollusks as possible indicators of multidrug-resistant Escherichia coli and other species of the Enterobacteriaceae family. Frontiers Mic. 2017;8:24.
75.
Capkin E, Terzi E, Altinok I. Occurrence of antibiotic resistance genes in culturable bacteria isolated from Turkish trout farms and their local aquatic environment. Dis Aquatic Organisms. 2015;114:127–137.
76.
Shen Y, Lv Z, Yang L, et al. Integrated aquaculture contributes to the transfer of mcr-1 between animals and humans via the aquaculture supply chain. Envir International. 2019;130:104708. https://doi.org/10.1016/j.envi....
77.
Lv L, Cao Y, Yu P, et al. Detection of mcr-1 gene among Escherichia coli isolates from farmed fish and characterization of mcr-1-bearing IncP plasmids. Antimicrobial Agents Chem. 2018;62:e02378–17. https://doi.org/10.1128/aac.02....
We process personal data collected when visiting the website. The function of obtaining information about users and their behavior is carried out by voluntarily entered information in forms and saving cookies in end devices. Data, including cookies, are used to provide services, improve the user experience and to analyze the traffic in accordance with the Privacy policy. Data are also collected and processed by Google Analytics tool (more).
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.