Online first
Current issue
Archive
Special Issues
About the Journal
Publication Ethics
Anti-Plagiarism system
Instructions for Authors
Instructions for Reviewers
Editorial Board
Editorial Office
Contact
Reviewers
All Reviewers
2025
2024
2023
2022
2021
2020
2019
2018
2017
2016
General Data Protection Regulation (RODO)
BRIEF COMMUNICATION
The Microorganism Detection System (SDM) for microbiological control of cosmetic products
1
Institute of Theory of Electrical Engineering, Measurement and Information Systems, University of Technology, Warsaw, Poland
2
NVT Limited Liability Company, Warsaw, Poland
3
Institute of Rural Health, Lublin, Poland
4
Department of Occupational Medicine, Medical University, Lublin, Poland
Corresponding author
Ann Agric Environ Med. 2021;28(4):705-708
KEYWORDS
artificial intelligencedetection of microorganismsimage interpretation algorithmmicrobiological safety
TOPICS
ABSTRACT
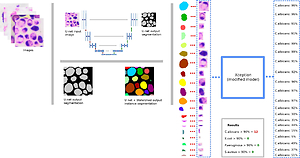
The Microorganism Detection System (SDM) is a new solution using artificial intelligence, unique on the international scale, to correctly identify and count microorganisms, with particular emphasis on specificlisted microorganisms (Document of Standard PN-EN ISO 17516–2014:11) – Candida albicans, Escherichia coli, Pseudomonas aeruginosa, Staphylococcus aureus. SDM enables the use of algorithms for microscopic image interpretation in the microbiological assessment of the cosmetics in accordance with the standard, providing an answer to whether the tested product complies with the standard. Apart from the software part of SDM, an integral part of the system is an innovative methodology for preparing a cosmetic sample for testing. The experiments confirm the high sensitivity and specificity of the SDM method, its repeatability and, above all, the comparability of the results with the methods of European standards.
ACKNOWLEDGEMENTS
The project was co-financed by the European Union from the European Regional Development Fund under the Intelligent Development programme. The project implemented under the competition 1 / 4.1.4 / 2018 / POIR: ‘Microorganism
detection system – SDM’, National Centre for Research and Development.
REFERENCES (17)
2.
Regulation (EC) No 1223/2009 of the European Parliament and of the Council of 30 November 2009 on cosmetic products. https://eur-lex.europa.eu/eli/... (access: 2021.11.07).
3.
Fukushima H, Katsube K, Hata Y, Kishi R, Fujiwara S. Rapid separation and concentration of food-borne pathogens in food samples prior to quantification by viable-cell counting and real-time PCR. Appl Environ Microbiol. 2007; 73(1): 92–100. doi: 10.1128/AEM.01772-06.
4.
Pertoft H. Fractionation of cells and subcellular particles with Percoll. J Biochem Biophys Methods. 2000; 44: 1–30. https://doi.org/10.1016/S0165-....
5.
Garbacz M, Malec A, Duda-Saternus S, Suchorab Z, Guz Ł, Łagód G. Methods for Early Detection of Microbiological Infestation of Buildings Based on Gas Sensor Technologies. Chemosensors. 2020; 8: 7. doi: 10.3390/chemosensors8010007.
6.
Bartholomew JW, Mittwer T. The Gram stain. Bacteriol Rev. 1952; 16(1): 1–29. doi: 10.1128/br.16.1.1-29.1952.
7.
Beveridge TJ. Use of the gram stain in microbiology. Biotech Histochem. 2001; 76(3): 111–8.
9.
Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation, CoRR: http://lmb.informatik.uni-frei... 2015 (access: 2021.11.07).
10.
Beucher S. The Watershed Transformation Applied to Image Segmentation, Scanning Microscopy, 1992, https://www.researchgate.net/p... (access: 2021.11.07).
11.
Bankhead P, Loughrey MB, Fernández JA, et al. QuPath. Open source software for digital pathology image analysis. Sci Rep. 2017; 7: 16878. doi.org/10.1038/s41598-017-17204-5.
12.
Redmon J, Farhadi A. YOLOv3: An Incremental Improvement, CoRR, 2018, https://arxiv.org/pdf/1804.027... (access: 2021.11.07).
13.
Chollet F. Xception: Deep Learning with Depthwise Separable Convolutionn, CoRR, 2017 https://arxiv.org/pdf/1610.023... (access: 2021.11.07).
15.
Alzubaidi L, Zhang J, Amjad I, et al. Review of deep learning: concepts, CNN architectures, challenges, applications, future directions. J Big Data. 2021; 8(1): 53. doi: 10.1186/s40537-021-00444-8.
16.
Shrestha YR, Krishna V, von Krogh G. Augmenting Organizational DecisionMaking with Deep Learning Algorithms: Principles, Promises, and Challenges. J Bus Res. 2021; 123: 588–603. https://doi.org/10.1016/j.jbus....
Share
RELATED ARTICLE
We process personal data collected when visiting the website. The function of obtaining information about users and their behavior is carried out by voluntarily entered information in forms and saving cookies in end devices. Data, including cookies, are used to provide services, improve the user experience and to analyze the traffic in accordance with the Privacy policy. Data are also collected and processed by Google Analytics tool (more).
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.