Online first
Current issue
Archive
Special Issues
About the Journal
Publication Ethics
Anti-Plagiarism system
Instructions for Authors
Instructions for Reviewers
Editorial Board
Editorial Office
Contact
Reviewers
All Reviewers
2025
2024
2023
2022
2021
2020
2019
2018
2017
2016
General Data Protection Regulation (RODO)
RESEARCH PAPER
Microbial community, pathogenic bacteria and high-risk anti-biotic resistance genes at two tropical coastal beaches adjacent to villages in Hainan, China
1
International School of Public Health and One Health, Hainan Medical University, China
2
Testing Institute of the Centre for Disease Control and Prevention, Hainan, China
These authors had equal contribution to this work
Ann Agric Environ Med. 2023;30(4):645-653
KEYWORDS
microbial communitycarbapenem-resistant Klebsiella pneumoniaeantimicrobial resistanceantibiotic-resistant genescoastal beachone health
TOPICS
ABSTRACT
Objective:
The aim of the study was to explore the correlation between characteristics of microbial community, pathogenic bacteria and high-risk antibiotic-resistant genes, between coastal beaches and a multi-warm-blooded host, as well as to determine potential species biomarkers for faecal source contamination on tropical coastal beaches in China.
Material and methods:
The ‘One-Health’ approach was used in a microbiological study of beaches and warm-blooded hosts. The microbial.community was analyzed using 16S rRNA gene amplicons and shotgun metagenomics on Illumina NovaSeq.
Results:
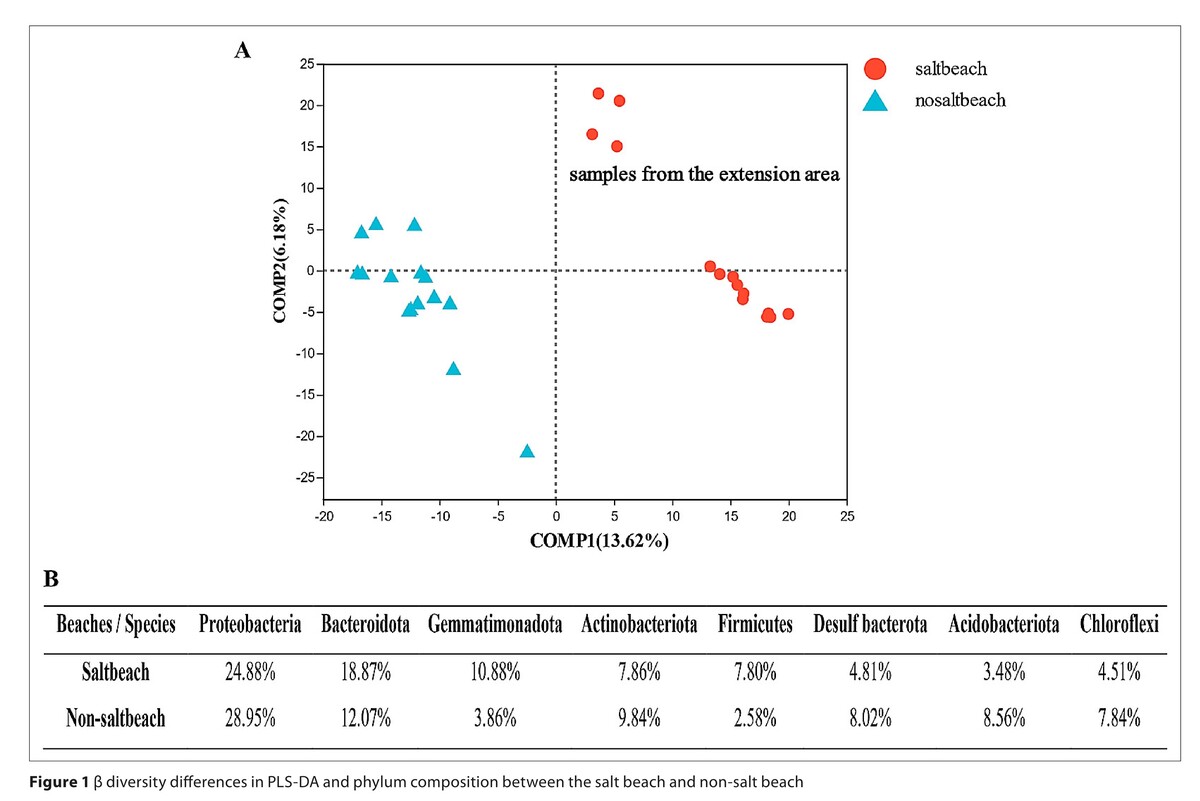
The Chao, Simpson, Shannon, and ACE indices of non-salt beach were greater than those of salt beaches at the genus and OTU levels (P < 0.001). Bacteroidota, Halanaerobiaeota, Cyanobacteria, and Firmicutes were abundant on salt beaches (P<0.01). Human-sourced microorganisms were more abundant on salt beaches, which accounted for 0.57%. Faecalibacterium prausnitzii and Eubacterium hallii were considered as reliable indicators for the contamination of human faeces. High-risk carbapenem-resistant Klebsiella pneumoniae and the genotypes KPC-14 and KPC-24 were observed on salt beaches. Tet(X3)/tet(X4) genes and four types of MCR genes co-occurred on beaches and humans; MCR9.1 accounted for the majority. Tet(X4) found among Cyanobacteria. Although rarely reported at Chinese beaches, pathogens, such as Vibrio vulnificus, Legionella pneumophila, and Helicobacter pylori, were observed.
Conclusions:
The low microbial community diversity, however, did not indicate a reduced risk. The transfer of high-risk ARGs to extreme coastal environments should be given sufficient attention.
The aim of the study was to explore the correlation between characteristics of microbial community, pathogenic bacteria and high-risk antibiotic-resistant genes, between coastal beaches and a multi-warm-blooded host, as well as to determine potential species biomarkers for faecal source contamination on tropical coastal beaches in China.
Material and methods:
The ‘One-Health’ approach was used in a microbiological study of beaches and warm-blooded hosts. The microbial.community was analyzed using 16S rRNA gene amplicons and shotgun metagenomics on Illumina NovaSeq.
Results:
The Chao, Simpson, Shannon, and ACE indices of non-salt beach were greater than those of salt beaches at the genus and OTU levels (P < 0.001). Bacteroidota, Halanaerobiaeota, Cyanobacteria, and Firmicutes were abundant on salt beaches (P<0.01). Human-sourced microorganisms were more abundant on salt beaches, which accounted for 0.57%. Faecalibacterium prausnitzii and Eubacterium hallii were considered as reliable indicators for the contamination of human faeces. High-risk carbapenem-resistant Klebsiella pneumoniae and the genotypes KPC-14 and KPC-24 were observed on salt beaches. Tet(X3)/tet(X4) genes and four types of MCR genes co-occurred on beaches and humans; MCR9.1 accounted for the majority. Tet(X4) found among Cyanobacteria. Although rarely reported at Chinese beaches, pathogens, such as Vibrio vulnificus, Legionella pneumophila, and Helicobacter pylori, were observed.
Conclusions:
The low microbial community diversity, however, did not indicate a reduced risk. The transfer of high-risk ARGs to extreme coastal environments should be given sufficient attention.
ACKNOWLEDGEMENTS
The research was funded by the Science and Technology
Department of Hainan Province (Grant No. ZDYF2020181),
and the National Natural Science Foundation of China (Grant
No. 81460487). The authors particularly express their thanks
to the participants in Danzhou County for their invaluable
collaboration and support.
REFERENCES (34)
1.
Moopantakath J, Imchen M, Kumavath R, Garcés E, Gasol JM, Ferrera I. Ubiquitousness of Haloferax and Carotenoid Producing Genes in Arabian Sea Coastal Biosystems of India. Marine drugs. 2021;19(8):442. https://doi.org/10.3390/md1908....
2.
Habibi, N, Uddin S, Lyons B, Al-Sarawi H A, Behbehani M, Shajan A, et al. Antibiotic Resistance Genes Associated with Marine Surface Sediments: A Baseline from the Shores of Kuwait. Sustainability. 2022;14:8029. https://doi.org/10.3390/su1413....
3.
Pulingam T, Parumasivam T, Gazzali AM, Sulaiman AM, Chee JY, Lakshmanan M, et al. Antimicrobial resistance: Prevalence, economic burden, mechanisms of resistance and strategies to overcome. Eur J Pharmaceutical Sci: Official J Eur Federation Pharmaceutical Sci. 2022;170:106103. https://doi.org/10.1016/j.ejps....
4.
Zhang Y, Zhao Z, Xu H, Wang L, Liu R, Jia X, et al. Fate of antibiotic resistance genes and bacteria in a coupled water-processing system with wastewater treatment plants and constructed wetlands in coastal eco-industrial parks. Ecotoxicol Environ Safety. 2023;252:114606.https://doi.org/10.1016/j.ecoe....
5.
Gorrasi S, Pasqualetti M, Franzetti A, Gonzalez-Martinez A, Gonzalez-Lopez J, Munoz-Palazon B, et al. Persistence of Enterobacteriaceae Drawn into a Marine Saltern (Saline di Tarquinia, Italy) from the Adjacent Coastal Zone. Water. 2021;13:1443. https://doi.org/10.3390/w13111....
6.
Saingam P, Di D, Yan T. Diversity and health risk potentials of the Enterococcus population in tropical coastal water impacted by Hurricane Lane. J Water Health. 2021;19(6):990–1001. https://doi.org/10.2166/wh.202....
7.
Gerken TJ, Roberts MC, Dykema P, Melly G, Lucas D, De LS, et al. Environmental Surveillance and Characterization of Antibiotic Resistant Staph-ylococcus aureus at Coastal Beaches and Rivers on the Island of Hawai’i. Antibiotics (Basel, Switzerland). 2021;10(8):980. https://doi.org/10.3390/antibi....
8.
Gambino D, Savoca D, Sucato A, Gargano V, Gentile A, Pantano L, et al. Occurrence of Antibiotic Resistance in the Mediterranean Sea. Antibi-otics. 2022;11(3):332. https://doi.org/10.3390/antibi....
9.
Su J, Fan J, Ming H, Guo G, Fu Y, Zhao X, et al. The Municipal Sewage Discharge May Impact the Dissemination of Antibi-otic-Resistant Escherichia coli in an Urban Coastal Beach. Water. 2022;14:1639. https://doi.org/10.3390/w14101....
10.
Harmon DE, Miranda OA, McCarley A, Eshaghian M, Carlson N, Ruiz C. Prevalence and characterization of carbapenem-resistant bacteria in water bodies in the Los Angeles-Southern California area. Microbiology Open. 2018;8(4):e00692. https://doi.org/10.1002/mbo3.6....
11.
Matamoros S, van Hattem JM, Arcilla Willemse N, Melles DC, Penders J, et al. Author Correction: Global phylogenetic analysis of Escherichia coli and plasmids carrying the mcr-1 gene indicates bacterial diversity but plasmid restriction. Sci Reports. 2020;10(1):2963. https://doi.org/10.1038/s41598....
12.
Shen Z, Hu Y, Sun Q, Hu F, Zhou H, Shu L, et al. Emerging Carriage of NDM-5 and MCR-1 in Escherichia coli From Healthy People in Multiple Regions in China: A Cross Sectional Observational Study. E Clin Med. 2018;6:11–20. https://doi.org/10.1016/j.ecli....
13.
Mohammad A, Narjess B, Hossein G, Mansoor K, Fariba A, Hadis F, et al. Colistin resistance mechanisms in Gram-negative bacteria: a Focus on Escherichia col. Letters in Applied Microbiol. 2023;76:1–13. https://doi.org/10.1093/lambio....
14.
Li R, Liu Z, Peng K, Liu Y, Xiao X, Wang Z. Co-occurrence of two tet(X) variants in an Empedobacter brevis of shrimp origin. Antimicrobial agents and chemotherapy, 2019;63(12):e01636–19. Advance online publication. https://doi.org/10.1128/AAC.01....
15.
He T, Wang R, Liu D, Walsh TR, Zhang RLY, et al. Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nature Microbiol. 2019:4(9):1450–1456. https://doi.org/10.1038/s41564....
16.
Chen C, Cui CY, Yu JJ, He Q, Wu XT, He YZ, et al. Genetic diversity and characteristics of high-level tigecycline resistance Tet(X) in Acinetobacter species. Genome medicine. 2020;12(1):111. https://doi.org/10.1186/s13073....
17.
WHO. (2022). One Health Joint Plan of Action launched to address health threats to humans, animals, plants and environment. World Health Organization.
18.
Yao Tang, Zhishu Liang, Guiying Li, Huijun Zhao, Taicheng An. Metagenomic profiles and health risks of pathogens and antibiotic resistance genes in various industrial wastewaters and the associated receiving surface water. Chemosphere. 2021;283:131224.
19.
Ren Y, Yu G, Shi C, Liu L, Guo, Han C, et al. Majorbio Cloud: A one-stop, comprehensive bioinformatic platform for multiomics analyses. iMeta 2022; doi:10.1002/IMT2.12.
20.
Shrivastava SR, Shrivastava PS, Ramasamy J, et al. World health organization releases global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. J Med Soc. 2018;32:76–77. doi:10.4103/JMS.JMS 25_17.
21.
Long WF, Li TJ, Yi GH, Liang F, Hu GY, Wu JZ, et al. Prevalence, phenotype and genotype characteristics of antibiotic resistance in coastal beach practitioners of tropical China. One Health Bulletin 2022;2(2):4–10. doi:10.4103/2773-0344.343630.
22.
Auladell A, Barberán A, Logares R, Garcés E, Gasol J M, Ferrera I, et al. Seasonal niche differentiation among closely related marine bacteria. ISME J. 2022;16(1):178–189. https://doi.org/10.1038/s41396....
23.
Byappanahalli MN, Nevers MB, Shively D, Nakatsu CH, Kinzelman JL, Phanikumar MS, et al. Influence of Filter Pore Size on Composition and Relative Abundance of Bacterial Communities and Select Host-Specific MST Markers in Coastal Waters of Southern Lake Michigan. Frontiers Microbiol. 2021;12, 665664. https://doi.org/10.3389/fmicb.....
24.
Ballesté E, Demeter K, Masterson B, Timoneda N, Sala-Comorera L, Meijer WG, et al. Implementation and integration of microbial source tracking in a river watershed monitoring plan. Sci Total Environ. 2020;736:139573. https://doi.org/10.1016/j.scit....
25.
Anton J, Oren A, Benlloch S, et al. Salinibacter ruber gen. nov., sp. nov., a novel, extremely halophilic member of the Bacteria from saltern crystallizer ponds. Int J Syst Evol Microbiol. 2002;52:485–491. https://doi.org/10.1099/002077....
26.
Zhang YX, Wang JH, Wu J. Antibiotic resistance genes might serve as new indicators for wastewater contami-nation of coastal waters: Spatial distribution and source apportionment of antibiotic resistance genes in a coastal bay. Ecological indicators. 2020;114:106299. doi:10.1016/j.ecolind.2020.106299.
27.
Li X, Harwood VJ, Nayak B, Weidhaas L. Ultrafiltration and Microarray for Detection of Microbial Source Tracking Marker and Pathogen Genes in Riverine and Marine Systems. Appl Environ Microbiol. 2016;82(5):1625–1635. https://doi.org/10.1128/AEM.02....
28.
Quaglia NC, Storelli MM, Scardocchia T, Lattanzi A, Celano GV, Monno R, et al. Helicobacter pylori: Survival in cultivable and non-cultivable form in artificially contaminated Mytilus galloprovincialis. Inter J Food Microbiol. 2020;312:108363. https://doi.org/10.1016/j.ijfo....
29.
Bridgemohan RSH, Deitch MJ, Ramsubhag A, Bridgemohan P, et al. Detection of Campylobacter jejuni Presence in Trinidad’s Aquatic En-vironments. Water, air, and soil pollution 2022;233:215. https://doi.org/10.1007/s11270....
30.
Zhang H, Wei W, Huang M, Umar Z, Feng Y. Erratum: Definition of a Family of Nonmobile Colistin Resistance (NMCR-1) Determinants Suggests Aquatic Reservoirs for MCR-4. Advanced science (Weinheim, Baden-Wurttemberg, Germany), 2020;7(17):2002530. https://doi.org/10.1002/advs.2....
31.
Shalygin SS, Kavulic K, Pietrasiak N, Bohunická M, Vaccarino MA, Chesarino NM, et al. Neotypification of Pleurocapsa fuliginosa and epitypification of P. minor (Pleurocapsales): resolving a polyphyletic cyanobacterial genus. Phytotaxa 2019;392(4):245–263. https://doi.org/10.11646/phyto....
32.
Umar Z, Chen Q, Tang B, Xu Y, Wang J, Zhang H, et al. The poultry pathogen Riemerella anatipestifer appears as a reservoir for Tet(X) tigecycline resistance. Environ Microbiol. 2021;23(12):7465–7482. https://doi.org/10.1111/1462-2....
33.
Zhang T, Cheng F, Yang H, Zhu B, Li C, Zhang YN, et al. Photochemical degradation pathways of cell-free antibiotic resistance genes in water under simulated sunlight irradiation: Experimental and quantum chemical studies. Chemosphere. 2022;302:134879. https://doi.org/10.1016/j.chem.... 2022.134879.
34.
Xu Y, You G, Yin J, Zhang M, Peng D, Xu J, et al. Salt tolerance evolution facilitates antibiotic resistome in soil microbiota: Evidences from dissemination evaluation, hosts identification and co-occurrence exploration. Environ Poll. (Barking, Essex: 1987), 2023;317:120830. https://doi.org/10.1016/j.envp....
Share
RELATED ARTICLE
We process personal data collected when visiting the website. The function of obtaining information about users and their behavior is carried out by voluntarily entered information in forms and saving cookies in end devices. Data, including cookies, are used to provide services, improve the user experience and to analyze the traffic in accordance with the Privacy policy. Data are also collected and processed by Google Analytics tool (more).
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.