Online first
Current issue
Archive
Special Issues
About the Journal
Publication Ethics
Anti-Plagiarism system
Instructions for Authors
Instructions for Reviewers
Editorial Board
Editorial Office
Contact
Reviewers
All Reviewers
2025
2024
2023
2022
2021
2020
2019
2018
2017
2016
General Data Protection Regulation (RODO)
RESEARCH PAPER
Analysis of the prevalence of colistin resistance among clinical strains of Klebsiella pneumoniae
1
Department of Laboratory Medicine; Chair of Microbiology, Immunology and Laboratory Medicine, Pomeranian
Medical University, Szczecin, Poland
2
Department of Diagnostic Immunology; Chair of Microbiology, Immunology and Laboratory Medicine, Pomeranian Medical University, Szczecin, Poland
3
Department of Clinical Microbiology, Chair of Microbiology, Immunology and Laboratory Medicine, Pomeranian
Medical University, Szczecin, Poland
4
Microbiological Laboratory, Independent Public Clinical Hospital No. 1, Szczecin, Poland
5
Microbiological Laboratory, Independent Public Clinical Hospital No. 2. Szczecin, Poland
Corresponding author
Agata Pruss
Department of Laboratory Medicine; Chair of Microbiology, Immunology and Laboratory Medicine; Pomeranian Medical University in Szczecin, Powstancow Wielkopolskich 72, 70-111, Szczecin, Poland
Department of Laboratory Medicine; Chair of Microbiology, Immunology and Laboratory Medicine; Pomeranian Medical University in Szczecin, Powstancow Wielkopolskich 72, 70-111, Szczecin, Poland
Ann Agric Environ Med. 2022;29(4):518-522
KEYWORDS
TOPICS
ABSTRACT
Introduction and objective:
Klebsiella pneumoniae is an essential component of the human gut microflora. However, it can pose a threat by causing opportunistic infections, especially in hospitalised or immunocompromised patients. It is a serious problem for health medicine, primarily because of increasing resistance to previously used antibiotics. Infections with multidrug-resistant strains are difficult to treat, creating a challenge for clinicians. Also of growing concern is the increasing resistance to the drug of last resort – colistin (CL). The aim of the study is to determine the prevalence of resistance to CL among clinical K. pneumoniae strains.
Material and methods:
The study was conducted on 200 clinical strains of K. pneumoniae. Drug susceptibility, production of resistance mechanisms, and determination of the minimum inhibitory concentration of CL were evaluated.
Results:
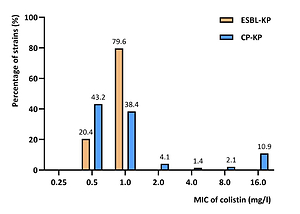
Of all isolates, 73.0% produced carbapenemases, while the remainder produced an extended substrate spectrum – β-lactamases (ESBLs). All strains showed a diverse antibiotic resistance profile. Resistance to CL was noted among 14.5% of carbapenemase-producing strains, particularly MBL and OXA-48. ESBL-positive strains showed full susceptibility to CL.
Conclusions:
Although a low rate of CL resistance was observed, this was true for strains simultaneously producing carbapenemases. Such strains should be under special epidemiological surveillance due to their potential to cause epidemic outbreaks. Monitoring the prevalence of clinical CL-resistant strains would allow for more effective counteraction against pathogens in various fields, including medicine, agriculture, veterinary medicine and industry.
Klebsiella pneumoniae is an essential component of the human gut microflora. However, it can pose a threat by causing opportunistic infections, especially in hospitalised or immunocompromised patients. It is a serious problem for health medicine, primarily because of increasing resistance to previously used antibiotics. Infections with multidrug-resistant strains are difficult to treat, creating a challenge for clinicians. Also of growing concern is the increasing resistance to the drug of last resort – colistin (CL). The aim of the study is to determine the prevalence of resistance to CL among clinical K. pneumoniae strains.
Material and methods:
The study was conducted on 200 clinical strains of K. pneumoniae. Drug susceptibility, production of resistance mechanisms, and determination of the minimum inhibitory concentration of CL were evaluated.
Results:
Of all isolates, 73.0% produced carbapenemases, while the remainder produced an extended substrate spectrum – β-lactamases (ESBLs). All strains showed a diverse antibiotic resistance profile. Resistance to CL was noted among 14.5% of carbapenemase-producing strains, particularly MBL and OXA-48. ESBL-positive strains showed full susceptibility to CL.
Conclusions:
Although a low rate of CL resistance was observed, this was true for strains simultaneously producing carbapenemases. Such strains should be under special epidemiological surveillance due to their potential to cause epidemic outbreaks. Monitoring the prevalence of clinical CL-resistant strains would allow for more effective counteraction against pathogens in various fields, including medicine, agriculture, veterinary medicine and industry.
REFERENCES (33)
1.
Podschun R, Ullmann U. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev. 1998;11(4):589–603.
2.
Schroll C, Barken KB, Krogfelt KA, et al. Role of type 1 and type 3 fimbriae in Klebsiella pneumoniae biofilm formation. BMC Microbiol. 2010 Jun 23;10:179.
3.
Paczosa MK, Mecsas J. Klebsiella pneumoniae: Going on the Offense with a Strong Defense. Microbiol Mol Biol Rev. 2016;80(3):629–661. doi: 10.1128/MMBR.00078-15.
4.
Lee CR, Lee JH, Park KS, et al. Antimicrobial Resistance of Hypervirulent Klebsiella pneumoniae: Epidemiology, Hypervirulence-Associated Determinants, and Resistance Mechanisms. Front Cell Infect Microbiol. 2017;7:483. doi: 10.3389/fcimb.2017.00483.
5.
Bush K, Bradford PA. Epidemiology of ß-Lactamase-Producing Pathogens. Clin Microbiol Rev. 2020;33(2):e00047–19. Published 2020 Feb 26. doi: 10.1128/CMR.00047-19.
6.
Fair RJ, Tor Y. Antibiotics and bacterial resistance in the 21st century. Perspect Medicin Chem. 2014;6:25–64. Published 2014 Aug 28. doi: 10.4137/PMC.S14459.
7.
Bialvaei AZ, Samadi Kafil H. Colistin, mechanisms and prevalence of resistance. Curr Med Res Opin. 2015 Apr;31(4):707–21. doi: 10.1185/03007995.2015.1018989.
8.
Stefaniuk EM, Tyski S. Colistin Resistance in Enterobacterales Strains – A Current View. Pol J Microbiol. 2019;68(4):417–427. doi: 10.33073/pjm-2019-055.
9.
El-Sayed Ahmed MAE, Zhong LL, Shen C, Yang Y, Doi Y, Tian GB. Colistin and its role in the Era of antibiotic resistance: an extended review (2000–2019). Emerg Microbes Infect. 2020;9(1):868–885. doi: 10.1080/22221751.2020.1754133.
10.
Nang SC, Morris FC, McDonald MJ, et al. Fitness cost of mcr-1-mediated polymyxin resistance in Klebsiella pneumoniae. J Antimicrob Chemother. 2018;73(6):1604–1610. doi: 10.1093/jac/dky061. PMID: 29514208.
11.
Olaitan AO, Morand S, Rolain JM. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol. 2014;5:643. doi: 10.3389/fmicb.2014.00643.
12.
Nang SC, Morris FC, McDonald MJ, et al. Fitness cost of mcr-1-mediated polymyxin resistance in Klebsiella pneumoniae. J Antimicrob Chemother. 2018;73(6):1604–1610. doi: 10.1093/jac/dky061.
13.
Liu YY, Wang Y, Walsh TR, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016 Feb;16(2):161–8. doi: 10.1016/S1473-3099(15)00424-7.
14.
Wang X, Wang Y, Zhou Y, et al. Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg Microbes Infect. 2018;7(1):122. doi: 10.1038/s41426-018-0124-z.
15.
EUCAST The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 9.0. 2019. http://www.eucast.org.
16.
Shon AS, Bajwa RP, Russo TA. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence. 2013 Feb 15;4(2):107–18. doi: 10.4161/viru.22718.
17.
Fils P, Cholley P, Gbaguidi-Haore H, et al. ESBL-producing Klebsiella pneumoniae in a University hospital: Molecular features, diffusion of epidemic clones and evaluation of cross-transmission. PLoS One. 2021;16(3). doi:10.1371/journal.pone.0247875.
18.
Saadatian Farivar A, Nowroozi J, Eslami G, et al. RAPD PCR Profile, Antibiotic Resistance, Prevalence of armA Gene, and Detection of KPC Enzyme in Klebsiella pneumoniae Isolates. Can J Infect Dis Med Microbiol. 2018;2018:6183162. doi: 10.1155/2018/6183162.
19.
Ocampo AM, Chen L, Cienfuegos A, et al. A Two-Year Surveillance in Five Colombian Tertiary Care Hospitals Reveals High Frequency of Non-CG258 Clones of Carbapenem-Resistant Klebsiella pneumoniae with Distinct Clinical Characteristics. Antimicrobial Agents Chemother. 2016; 60(1): 332–342. doi: 10.1128/AAC.01775-15.
20.
Xercavins M, Jiménez E, Padilla E, et al. High clonal diversity of ESBL-producing Klebsiella pneumoniae isolates from clinical samples in a non-outbreak situation. A cohort study. Antimicrob Resist Infect Control. 2020;9(1):5. doi: 10.1186/s13756-019-0661-9.
21.
Eftekhar F, Nouri P. Correlation of RAPD-PCR Profiles with ESBL Production in Clinical Isolates of Klebsiella pneumoniae in Tehran. J Clin Diagn Res. 2015;9(1):DC01-DC3. doi: 10.7860/JCDR/2015/10651.5373.
22.
Unlu O, Demirci M. Detection of carbapenem-resistant Klebsiella pneumoniae strains harboring carbapenemase, beta-lactamase and quinolone resistance genes in intensive care unit patients. GMS Hyg Infect Control. 2020;15:Doc31. doi: 10.3205/dgkh000366.
23.
Zhang X, Chen D, Xu G, et al. Molecular epidemiology and drug resistant mechanism in carbapenem-resistant Klebsiella pneumoniae isolated from pediatric patients in Shanghai, China. PLoS One. 2018;13(3):e0194000. doi: 10.1371/journal.pone.0194000.
24.
Calia C, Pazzani C, Oliva M, et al. Carbapenemases-producing Klebsiella pneumoniae in hospitals of two regions of Southern Italy. APMIS. 2017 May;125(5):491–498. doi: 10.1111/apm.12666.
25.
Petrosillo N, Taglietti F, Granata G. Treatment options for colistin resistant Klebsiella pneumoniae: present and future. J Clin Med. 2019; 8:934. doi: 10.3390/jcm8070934.
26.
Pena I, Picazo JJ, Rodríguez-Avial C, et al. Carbapenemase-producing Enterobacteriaceae in a tertiary hospital in Madrid, Spain: high percentage of colistin resistance among VIM-1-producing Klebsiella pneumoniae ST11 isolates. Int J Antimicrob Agents. 2014 May;43(5):460–4. doi: 10.1016/j.ijantimicag.2014.01.021.
27.
Capone A, Giannella M, Fortini D, et al. High rate of colistin resistance among patients with carbapenem-resistant Klebsiella pneumoniae infection accounts for an excess of mortality. Clin Microbiol Infect. 2013 Jan;19(1):E23-E30. doi: 10.1111/1469-0691.12070.
28.
Rojas LJ, Salim M, Cober E, et al. Colistin Resistance in Carbapenem-Resistant Klebsiella pneumoniae: Laboratory Detection and Impact on Mortality. Clin Infect Dis. 2017 Mar 15;64(6):711–718. doi: 10.1093/cid/ciw805.
29.
Can F, Menekse S, Ispir P, et al. Impact of the ST101 clone on fatality among patients with colistin-resistant Klebsiella pneumoniae infection. J Antimicrob Chemother. 2018 May 1;73(5):1235–1241. doi: 10.1093/jac/dkx532.
30.
Jayol A, Poirel L, Dortet L, et al. National survey of colistin resistance among carbapenemase-producing Enterobacteriaceae and outbreak caused by colistin-resistant OXA-48-producing Klebsiella pneumoniae, France, 2014. Euro Surveill. 2016;21(37):30339. doi: 10.2807/1560-7917.ES.2016.21.37.30339.
31.
Jafari Z, Ali Harati A, Haeili M, et al. Molecular Epidemiology and Drug Resistance Pattern of Carbapenem-Resistant Klebsiella pneumoniae Isolates from Iran. Microbial Drug Resistance. 2019;25(3):336–343. doi: 10.1089/mdr.2017.0404.
32.
Baraniak A, Grabowska A, Izdebski R, et al. Molecular characteristics of KPC-producing Enterobacteriaceae at the early stage of their dissemination in Poland, 2008–2009. Antimicrob Agents Chemother. 2011;55(12):5493–9. doi: 10.1128/AAC.05118-11.
33.
Sękowska A, Chudy M, Gospodarek-Komkowska E. Emergence of colistin-resistant Klebsiella pneumoniae in Poland. Acta Microbiol Immunol Hung. 2019;67(1):18–22. doi: 10.1556/030.66.2019.028.
We process personal data collected when visiting the website. The function of obtaining information about users and their behavior is carried out by voluntarily entered information in forms and saving cookies in end devices. Data, including cookies, are used to provide services, improve the user experience and to analyze the traffic in accordance with the Privacy policy. Data are also collected and processed by Google Analytics tool (more).
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.